
When the run has completed take some time to review the output files. So now issue a run command to actually find the hydrogen bonds. That is because CPPTRAJ needs to actually process trajectories before it can find the hydrogen bonds. Note that at this point although we have set up hydrogen bond commands, there isn’t actually any hydrogen bond data yet. create nhbvtime.agr All Backbone All Allįinally, we will add an RMSD command to calculate backbone RMSD to the first frame as a means to track unfolding ( Note this is not part of hbond analysis): rms BBrmsd out BBrmsd.agr Number of solute-solvent hydrogen bonds (aspect: ) and number of solute-solvent-solute bridges (aspect: ). We will now issue a command that will create a file containing only the number of solute-solute hydrogen bonds (aspect: ) from both commands, as well as
Protein backbone hydrogen bonds series#
The uuseries keyword will write the raw hydrogen bond time series data to bbhbond.gnu, which can be visualized using gnuplot. hbond Backbone avgout BB.avg.dat series uuseries bbhbond.gnu We will also specify the series keyword so that the time series of each found hydrogen bond is recorded for further analysis (1 for hydrogen bond present, 0 for not present). As such we will provide a mask so that only C, O, N, and H atoms will be considered as potential donors/acceptors. The second hbond command will be used to track only solute-solute backbone hydrogen bonds. Average statistics on each found hydrogen bond will be found in the files ‘All.UU.avg.dat’, ‘All.UV.avg.dat’, and ‘’. The number of solute-solute and solute-solvent hydrogen bonds, as well as the number of bridges and the identity of bridging residues will be written to ‘’. The first hbond command will be used to track all solute-solute and solute-solvent hydrogen bonds (all solvent residues are named WAT, the Amber default), as well as solute-solvent-solute bridging interactions: hbond All out solventdonor :WAT solventacceptor \Īvgout All.UU.avg.dat solvout All.UV.avg.dat bridgeout Load the topology and trajectory with the following commands: parm 7 Performing Hydrogen Bond Analysis with CPPTRAJ Note that although input is provided in a file, users are encouraged to use the interactive mode to become better familiar with CPPTRAJ workflow and command options. : MD trajectory, unfolding of Trpzip2 at 345 K.In this example we will use CPPTRAJ to find and track hydrogen bonds during the unfolding of the model beta hairpin peptide Trpzip2 ( PDB 1LE1) at 345 K. Amber, Charmm) hydrogen bond interactions arise solely from non-bonded terms and therefore their strength is not dependent on angle. However in most pairwise additive force fields (e.g. Note that in nature, the strength of a hydrogen bond is determined by both distance and angle (having to do with optimal overlap of molecular orbitals). two or more solute residues hydrogen bonded to a single solvent residue).
CPPTRAJ will also track solute to solvent hydrogen bonds and solvent bridging interactions (i.e.
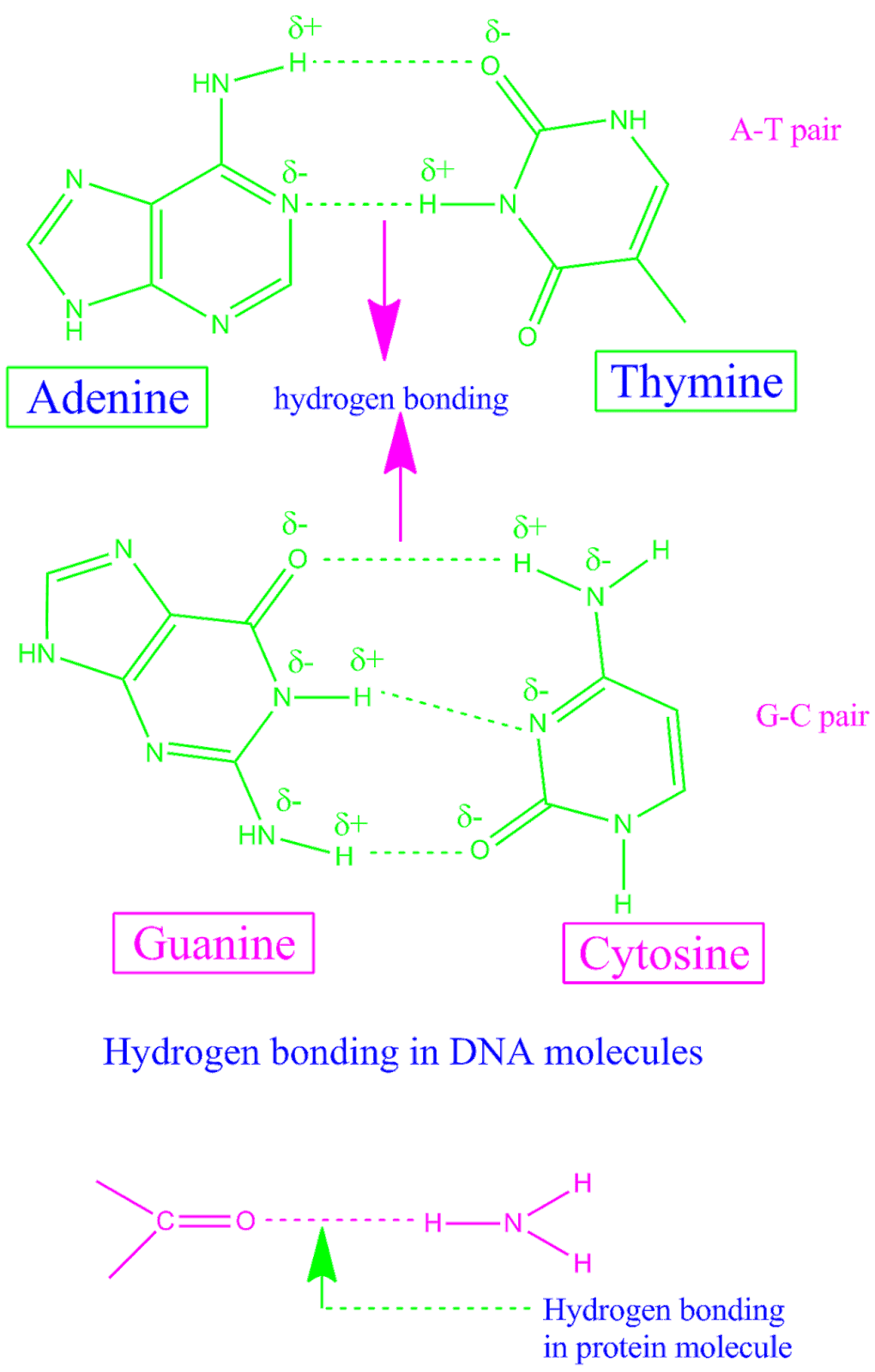
Hydrogen bond donors and acceptors can either be searched for automatically (using the FON criterion) or can be explicitly defined by the user via atom masks. Hydrogen bonds are determined using simple geometric criteria: the donor to acceptor heavy atom distance, and optionally the donor-hydrogen-acceptor angle. hydrogens bonded to F, O, and N atoms can be donated, and F, O, and N atoms can be acceptors (although there are exceptions).ĬPPTRAJ will find and track hydrogen bonds over the course of a trajectory. The general rule of thumb for determining whether atoms can hydrogen bond is “hydrogen bonds are FON”, i.e. Both the donor and acceptor atoms are typically quite electronegative. They are formed when a single hydrogen atom is effectively shared between the heavy atom it is covalently bonded to (the hydrogen bond donor) and another heavy atom (the hydrogen bond acceptor). Hydrogen bonds are an important non-covalent structural force (primarily electrostatic in nature) in molecular systems. between a protein and a small molecule) follow this recipe. For an example about hydrogen bonds between two independent systems (i. This recipe focuses on the analysis of intra-molecular hydrogen bonds as a protein unfolds at high temperature.

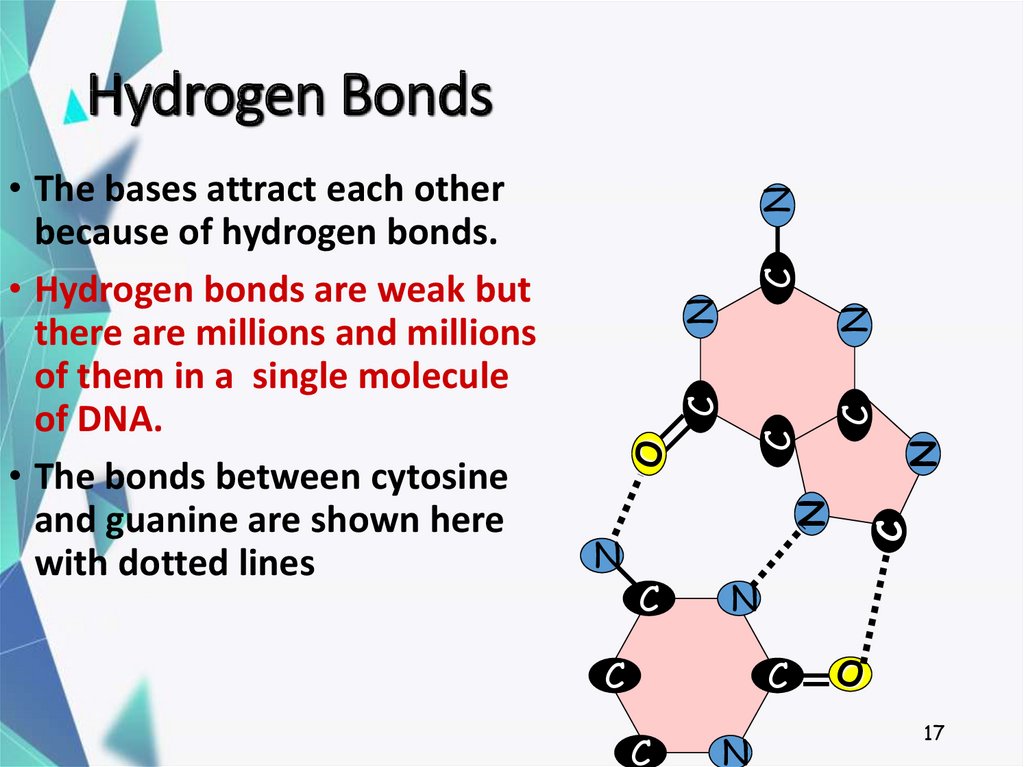
You want to study the hydrogen-bonds within a protein system


 0 kommentar(er)
0 kommentar(er)
